Research
Research
Inhibitors against serine β-lactamases
Structure-based development of non-β-lactam inhibitors against serine β-lactamases
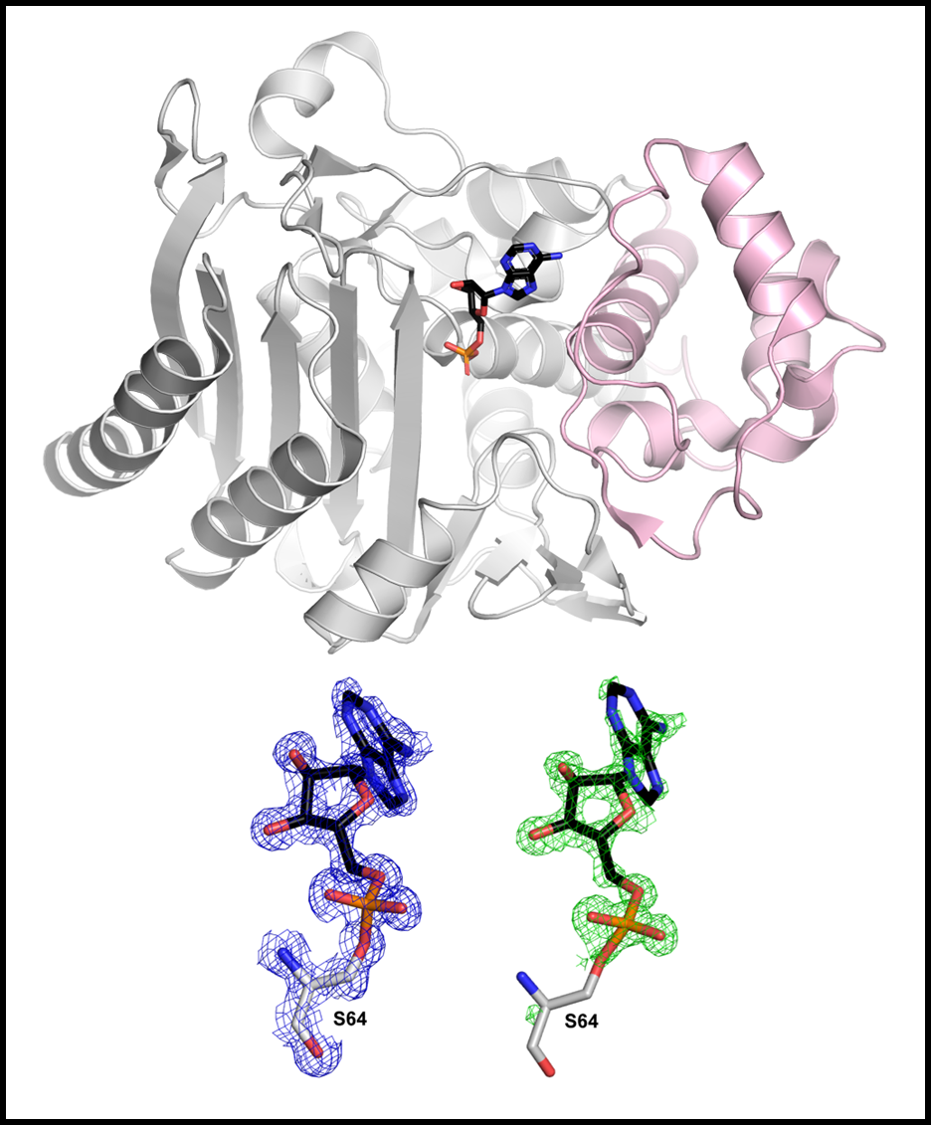
Class C β-lactamase
& antibiotics candidate
β-Lactams (penicillins, cephalosporins, and carbapenems) are the most widely used antibiotics to treat bacterial infections. Clinical application of β-lactams has been accompanied by the emergence of bacterial resistance to these antibiotics, which is a great threat to public health. Expression of β-lactamases is a prevalent resistance mechanism of bacteria to β-lactams. β-Lactamases inactivate β-lactams by hydrolyzing the amide bond in the β-lactam ring, the core structure of β-lactams. They are grouped into four classes, A, B, C and D, on the basis of sequence homology among which class A, B, and D are serine β-lactamases and metallo-β-lactamases belong to class B. Although class C β-lactamases along with class A enzymes are the most commonly encountered of the four classes in clinics, class C β-lactamases are more problematic than class A enzymes. Class C β-lactamases can confer resistance to cephamycins (cefoxitin and cefotetan), penicillins, and cephalosporins and are not significantly inhibited by clinically used β-lactamase inhibitors. In contrast, class A β-lactamases are not able to confer resistance to cephamycins and the enzymes are generally susceptible to inhibition by clinically-used inhibitors.
The development of inhibitors against β-lactamases is an effective strategy to cope with the β-lactamase-mediated antibiotic resistance since β-lactams maintain their antibiotic activity in the presence of inhibitors that block β-lactamases. In fact, three classical β-lactam inhibitors (clavulanate, sulbactam, and tazobactam) sharing the β-lactam backbone are clinically used in combination with β-lactam antibiotics (e.g. amoxicillin-clavulanate, ticarcillin-clavulanate, ampicillin-sulbactam , piperacillin-tazobactam, and cefoperazone-sulbactam). These clinical inhibitors are especially active on class A enzymes, displaying much less or no effect on other β-lactamaases. Furthermore, β-lactamases resistant to the β-lactam inhibitors are emerging, which highlights the need to develop non-β-lactam inhibitors with broad-spectrum efficacy.
Our research team has discovered non-β-lactam inhibitors against serine β-lactamases from cellular metabolites. The inhibitors are most effective against class C β-lactamases with extended substrate spectrum. The chemical structures of these novel inhibitors are different from those of existing inhibitors. Remarkably, one of them exhibited an in vivo efficacy without side-effects in mouse models. One concern regarding these inhibitors is their low affinity toward target enzymes, which is the cause of high dose for efficacy in vivo. To overcome the low affinity through designing potent inhibitors, we will reveal the crystal structures of the complexes between β-lactamases and these inhibitors, which will provide an invaluable platform to modify the structure of these novel inhibitors and also to understand their inhibition mechanisms. Our final goal is to develop non-β-lactam inhibitors that are effective towards all serine β-lactamases.
[Related publications]
- Bo-Gyeong Jeong¶, Myeong-Yeon Kim¶, Chang-Sook Jeong¶, Hackwon Do, Jisub Hwang, Jun Hyuck Lee*, and Sun-Shin Cha*
"Characterization of the extended substrate spectrum of the class A β-lactamase CESS-1 from Stenotrophomonas sp. and structure-based investigation into its substrate preference"
- International Journal of Antimicrobial Agents, Apr 6;107171(2024)
- International Journal of Antimicrobial Agents, Apr 6;107171
- Bo-Gyeong Jeong¶, Myeong-Yeon Kim¶, Chang-Sook Jeong¶, Hackwon Do, Jisub Hwang, Jun Hyuck Lee*, and Sun-Shin Cha*
Ji-Min Woo¶, Myeong-Yeon Kim¶, Ji-Won Song¶, Yoonjin Baeg, Hye-Jin Jo, Sun-Shin Cha*, Jin-Byung Park*
"Engineering of a bacterial outer membrane vesicle to a nano-scale reactor for the biodegradation of β-lactam antibiotics"
Journal of Biotechnology, Sep 10;356:1-7 (2022)
Yunseok Heo, Soo-Bong Park, Ye-Eun Jeon, Ji-Hye Yun, Bo-Gyeong Jeong, Sun-Shin Cha*, Weontae Lee*
"Structural and functional identification of uncharacterized metallo-β-lactamase superfamily protein TW9814 as a phosphodiesterase with unique metal coordination"
Acta Crystallogr D structural Biology, Apr 1;78(Pt 4):532-541(2022)
Bo-Gyeong Jeong, Jung-Hyun Na, Da-Woon Bae, Soo-Bong Park, Hyi-Seung Lee*, Sun-Shin Cha*
"Crystal structure of AmpC BER and molecular docking lead to the discovery of broad inhibition activities of halisulfates against β-lactamases"
Computational and Structural Biotechnology Journal, Dec 17;19:145-152 (2020)
Da-Woon Bae, Ye-Eun Jung, Bo-Gyeong Jeong, Sun-Shin Cha*
"Novel inhibition mechanism of carbapenems on the ACC-1 class C β-lactamase"
Archives of Biochemistry and Biophysics, Oct 30;693:108570 (2020)
Da-Woon Bae, Ye-Eun Jung, Young Jun An, Jung-Hyun Na, Sun-Shin Cha*
"Structural insights into catalytic relevances of substrate poses in ACC-1"
Antimicrobial Agents and Chemotherapy, Oct;63(11):e01411-19 (2019)
Jung-Hyun Na, Tae Hee Lee, Soo-Bong Park, Min-Kyu Kim, Bo-Gyeong Jeong, Kyung Min Chung*, Sun-Shin Cha*
"In vitro and In vivo Inhibitory Activity of NADPH against the AmpC BER Class C β-Lactamase"
Frontiers in Cellular and Infection Microbiology, 8:441 (2018)
Jung-Hyun Na, Young Jun An and Sun-Shin Cha*
"GMP and IMP are competitive inhibitors of CMY-10, an extended-spectrum class C β-lactamase"
Antimicrobial Agents and Chemotherapy, 61:5,e00098-17 (2017)
Min-Kyu Kim, Young Jun An, Jung-Hyun Na, Jae-Hee Seol, Ju Yeon Ryu,Jin-Won Lee, Lin-Woo Kang, Kyung Min Chung, Jung-Hyun Lee, Jeong Hee Moon, Jong Seok Lee, and Sun-Shin Cha*
"Structural and mechanistic insights into the inhibition of class C β-lactamases through the adenylylation of the nucleophilic serine"
The Journal of Antimicrobial Chemotherapy, 72 (3): 735-743 (2017)
Sun-Shin Cha*and Young Jun An
"Crystal structure of EstSRT1, a family VIII carboxylesterase displaying hydrolytic activity toward oxyimino cephalosporins"
Biochemical and Biophysical Research Communications, Sep 16;478(2):818-824 (2016)
Jung-Hyun Na and Sun-Shin Cha*
"Structural basis for the extended substrate spectrum of AmpC BER and structure-guided discovery of the inhibition activity of citrate against the class C β-lactamases AmpC BER and CMY-10"
Acta Crystallogr D Structural Biology, D72, 976-985 (2016)
Sun-Shin Cha*, Young Jun An, Chang-Sook Jeong,Min-Kyu Kim, Jeong Ho Jeon, Chang-Muk Lee, Hyun Sook Lee, Sung Gyun Kang, and Jung-Hyun Lee
"Structural basis for the β-lactamase activity of EstU1, a family VIII carboxylesterase"
Proteins: Structure, Function, and Bioinformatics, Nov;81(11):2045-2051 (2013)
Jae Young Kim, Ha Il Jung, Young Jun An, Jung Hun Lee, So Jung Kim, Seok Hoon Jeong, Kye Joon Lee, Pann-Ghill Suh, Heung-Soo Lee, Sang Hee Lee* and Sun-Shin Cha*
"Structural basis for the extended substrate spectrum of CMY-10, a plasmid-encoded class C β-lactamase"
Molecular Microbiology, May;60(4):907-916 (2006)
Jae Seok Song, Seon Ju Jang, Jae Jin Lee, Jung Hun Lee, Il Kwon Bae, Byeong Chul Jeong, Sun-Shin Cha, Jung-Hyun Lee, Soon-Kwang Hong and Sang Hee Lee*
"Association of blaCMY-10 gene with a novel complex class 1 integron carrying an ISCR1 element in clinical isolates from Korea"
Clinical Microbiology and Infection, Jul; 16:1013-1017 (2010)
[Opinions & Correspondences]
-Sang Hee Lee, Seok Hoon Jeong, Sun-Shin Cha
"Strategies to minimise antibiotic resistance"
The Lancet Infectious Diseases, Nov;5(11):668-670 (2005)
-Sang Hee Lee, Seok Hoon Jeong, Sun-Shin Cha
"Screening for carbapenem-resistant Gram-negative bacteria"
The Lancet Infectious Diseases, Nov;6(11):662-684 (2006)
-Sang Hee Lee, Jung Hun Lee, Myong Jin Heo, Il Kwon Bae, Seok Hoon Jeong, Sun-Shin Cha
"Exact location of the region responsible for the extended substrate spectrum in class C β-lactamases"
Antimicrob Agents Chemother., Oct;51(10): 3778-3779 (2007)
-Jung Hun Lee, Seok Hoon Jeong, Sun-Shin Cha, Sang-Hee Lee
"A lack of drugs for antibiotic resistant Gram-negative bacteria"
Nature Reviews Drug Discovery, Nov;6, 938-939 (2007)
-Jung Hun Lee, Seok Hoon Jeong, Sun-Shin Cha, Sang Hee Lee
"New disturbing trend in antimicrobial resistance of Gram-negative pathogens"
PLoS Pathogens, Mar;5(3):e1000221 (2009)

